PUBLICATIONS
 We are thrilled to announce a significant milestone in the field of glyco-immunopeptidomics achieved with the leadning contribution of ICCVS team. In a recent publication in Nature Communications, Georges Bedran (the 1st author of the paper), our PhD student together with his supervisor, Dr. Javier Alfaro (ICCVS) and in collaboration with bioinformatician team from University of Michigan Medical School (UMICH) led by prof. Alexey I. Nesvizhskii (the corresponding author) unveiled an innovative computational tool and resource for studying glycosylated MHC-associated peptides (MAPs). This study sheds light on the crucial role of these peptides and their presentation via MHC class II to the immune system.
We are thrilled to announce a significant milestone in the field of glyco-immunopeptidomics achieved with the leadning contribution of ICCVS team. In a recent publication in Nature Communications, Georges Bedran (the 1st author of the paper), our PhD student together with his supervisor, Dr. Javier Alfaro (ICCVS) and in collaboration with bioinformatician team from University of Michigan Medical School (UMICH) led by prof. Alexey I. Nesvizhskii (the corresponding author) unveiled an innovative computational tool and resource for studying glycosylated MHC-associated peptides (MAPs). This study sheds light on the crucial role of these peptides and their presentation via MHC class II to the immune system.
In this study we embarked on an exploration of the intricate landscape of post-translational modifications found on MAPs. Harnessing collective expertise of the ICCVS and prof. Nesvizhskii’s team (UMICH), we advanced our understanding of glycosylated MAPs. This collaboration has led to the development of an advanced computational tool that addresses the critical requirement for rapid and precise identification of non-enriched glycosylated MAPs from mass spectrometry data.
The current study resulted in the creation of a comprehensive resource known as HLA-Glyco. This resource contains over 3,400 HLA class II N-glycopeptides originating from 1,049 protein-glycosylation sites which are freely available online in accordance with open science idea (https://hla-glyco.nesvilab.org) and facilitates further research and exploration in the field of glyco-immunopeptidomics. Through an in-depth examination of HLA-Glyco, the team made intriguing discoveries regarding the characteristics and patterns of glycosylated MAPs. Notably, they revealed insights into truncated glycans, conserved HLA-binding cores, and the positional specificity of glycosylation. These findings contribute to a deeper understanding of how glycosylation influences antigen presentation and the immune response.
These findings have significant implications for enhancing our understanding of how glycosylation influences antigen presentation and is of great importance for anti-cancer vaccines development and generation of immune activation strategies. If you wish to delve deeper into this remarkable paper, it can be accessed here:
Bedran, G, Polasky, DA, Hsiao, Y, Yu F, da Veiga Leprevost, F, Alfaro, JA, Cieslik, M, Nesvizhskii, Alexey I. Unraveling the glycosylated immunopeptidome with HLA-Glyco. Nature Communications 14, 3461 (2023). https://doi.org/10.1038/s41467-023-39270-2
Current trends in immunology are progressing towards single-cell analyses of transcriptomes and proteomes. High-throughput and highly sensitive protein identification/sequencingis still just a dream, but it would lead to  awave of transformative biological/biomedical applications across fields akin to what was seen in the genomic revolution. Hence, why the journal Nature has placed single-molecule protein sequencing as a major technology to watch this year. In a series of 3 perspective pieces (2014, 2021, 2023) in Nature Methods involving over 40 researchers worldwide, dr Javier Alfaro from the International Centre for Cancer Vaccine Science at the University of Gdańsk and his colleagues have progressively identified a problem in how we infer proteomes today. He has identified a field of research that is bridging technologies from the genomics community to the proteomics community, and has pointed this community towards the technical issues they must overcome to sequence the proteome. Dr Alfaro is now starting a series of low-cell number proteomics studies in Ovarian Cancer with the University of Victoria (Canada), and would like to leverage his developed network through these papers towards spatial transcriptomics and proteomics technologies, other spatial technologies like mass-spec imaging to better characterize the immune-cancer/immune-disease synapse. Of course new and upcoming technologies still feel far away, but you never know how quickly technical and computational hurdles will be overcome as Javier and his colleagues have described in a communication.
awave of transformative biological/biomedical applications across fields akin to what was seen in the genomic revolution. Hence, why the journal Nature has placed single-molecule protein sequencing as a major technology to watch this year. In a series of 3 perspective pieces (2014, 2021, 2023) in Nature Methods involving over 40 researchers worldwide, dr Javier Alfaro from the International Centre for Cancer Vaccine Science at the University of Gdańsk and his colleagues have progressively identified a problem in how we infer proteomes today. He has identified a field of research that is bridging technologies from the genomics community to the proteomics community, and has pointed this community towards the technical issues they must overcome to sequence the proteome. Dr Alfaro is now starting a series of low-cell number proteomics studies in Ovarian Cancer with the University of Victoria (Canada), and would like to leverage his developed network through these papers towards spatial transcriptomics and proteomics technologies, other spatial technologies like mass-spec imaging to better characterize the immune-cancer/immune-disease synapse. Of course new and upcoming technologies still feel far away, but you never know how quickly technical and computational hurdles will be overcome as Javier and his colleagues have described in a communication.
Read the publication: MacCoss, M.J., Alfaro, J.A., Faivre, D.A. et al. Sampling the proteome by emerging single-molecule and mass spectrometry methods. Nat Methods 20, 339–346 (2023). https://doi.org/10.1038/s41592-023-01802-5
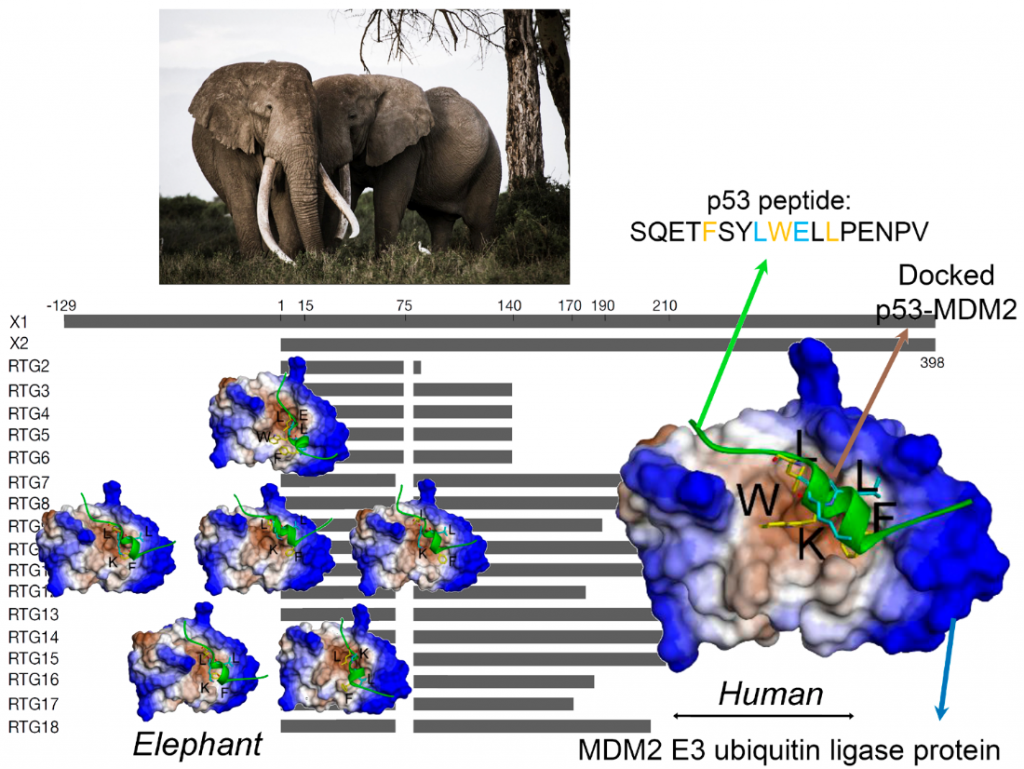
Elephants found ways to avoid Cancer – Article to come out in Molecular Biology and Evolution (IF: 16.2): https://academimc.oup.com/mbe/advance-article/doi/10.1093/molbev/msac149/6632613
Every day our cells replicate. For this the genes in each cell are copied by the process of DNA replication. Ideally, all new cells are exact copies of the older ones. However, there can be issues and the proteins mediating the replication of the DNA make mistakes and mutate. Not often but it adds up during the millions of daily gene activities. Most of these mistakes are repaired by the cell immediately. However, the number of mutations and the quality of repairs is affected by both genetic and external/living circumstances. Toxic compounds, stress, poor living conditions and ageing increase the mutation rate, sometimes massively.
Cancers are caused by accumulations of such gene mutations and over time, the risk of cancer increases. The longer we live the larger this risk. Spoiler alert: elephants are much larger than we and reach comparable ages yet they rarely develop cancers. Now a new study of the elephant genome conducted by researchers from International Centre for Cancer Vaccine Science, University of Gdańsk (Monikaben Padariya, Umesh Kalathiya, Ted Hupp, Robin Fåhraeus,) in collaboration with UK- Institute of Genetics and Cancer, University of Edinburgh (Mia-Lyn Jooste,Ted Hupp) and Department of Zoology, University of Oxford (Fritz Vollrath), France -Inserm UMRS1131, Institut de Génétique Moléculaire, France (Robin Fåhraeus and Konstantinos Karakostis), Czech Republic- Research Centre for Applied Molecular Oncology, Masaryk Memorial Cancer Institute (Borek Vojtesek, Robin Fåhraeus), Department of Medical Biosciences, Umeå University, Sweden (Robin Fåhraeus), Spain- Institut de Biotecnologia i de Biomedicina, Universitat Autònoma de Barcelona (and Konstantinos Karakostis) and Kenya- Save the Elephants Marula Manor, Nairobi (Fritz Vollrath) provides clues to important details in the process of cell health and sustainable tissue ageing.
P53 is a key player regulating the repairing mechanisms of the DNA and thus acts as a powerful suppressor of uncontrolled cell growth i.e. tumours. Hence P53 is often called the ‘guardian of the genome’. The p53 protein is activated when DNA is damaged and helps the cell orchestrate a response that pauses DNA replication and to repair any uncorrected copies. In normal conditions, p53 is inactivated by another protein, the oncogene MDM2 E3 ubiquitin ligase. In effect, the correct p53 – MDM2 handshaking permits healthy cells to divide and replicate but when the DNA is damaged, p53 is relieved of MDM2 and can support the DNA damage respοnse and repair. If the repair fails or the damage is extensive, then p53 takes the process one step further and programs the cell to self-destruct i.e. die. This so-called apoptosis prevents any mutations bad for cell health to be further replicated and form tumour tissues.
Back to the Spoiler alert: How can elephants help us to study critical phases in the p53-MDM2 handshaking procedure? Recent studies, have shown that the elephant has 40 copies of the p53 gene i.e. 20 genes with 2 alleles each. This discovery is very exciting indeed, as all other creatures seem to have only one gene i.e. 2 copies for p53. Thus, in effect, the elephant seems genetically over-endowed with a protein known to provide protection against cancers. Not only that, but each of the 40 copies can be slightly different (isoform) from the others, giving an elephant individual a much wider range of molecular anti-cancer interactions than a human would have with his/her two copies.
Now, in the current study our scientists have used a pioneer bioinformatic models to investigate details of the molecular interactions i.e. the handshaking between MDM2 and p53. By combining biochemical analysis with computer simulations, the team was able to show a remarkable effect. Because of minor variations in their molecular sequence, the different p53 isoforms have different molecular structures. It turns out that these structurally small differences which can have functionally very large effects with the altered 3-dimensional shapes of the p53 isoforms seriously affecting their handshake with MDM2. As a result, some of these p53 isoforms are able to escape the degradation mediated by MDM2.
First author of the publication, Dr. Monikaben Padariya from International Centre for Cancer Vaccine Science University of Gdańsk, says: “Alongside in vitro assays, the in silico molecular docking simulations provided complementary molecular level details for the p53-MDM2 systems. Our findings explored the FxxxWxxL motif from the BOX-I of p53. Such motifs increase the binding capacity to MDM2 and mutations within it induce changes on the conformation, as well as the positioning of the p53 BOX-I motif over the MDM2 interphase”.
Corresponding author Dr. Umesh Kalathiya from International Centre for Cancer Vaccine Science, University of Gdańsky adds: “Computational structural biology techniques often convey crucial fundamental information, that guides to gain insight into the molecular principles underlying biological processes. For the first time, molecular simulations were used to explore functional protein-protein intermolecular aspects of elephant (Loxodonta africana) p53 isoforms with MDM2”.
Crucially, this new research has identified how the 20 different molecules in this amazing population of p53 molecules are activated and how this can lead to an increased sensitivity and response against carcinogenic conditions. While this study is only one small step in our fight against cancer it shows how elephants have evolved defenses that provide us with powerful new insights and novel tools to study the mechanics of cancer.
“This is an existing development for our understanding of how p53 contributes to prevent cancer development. In humans, the same p53 protein is responsible for deciding if cells should stop proliferating or go into apoptosis but how p53 makes this decision has been difficult to elucidate. The existence of several p53 isoforms in elephants with different capacities to interact with MDM2 offers an existing new approach to shed new light on p53’s tumour suppressor activity”- says prof. Robin Fahraeus from ICCVS and Inserm UMRS1131, Paris, one of the authors.
This research establishes the elephant as a natural model to study the structural characteristics of the molecular interactions between the p53 tumour suppressor and its main regulator, MDM2; and set the mechanistic basis for the development of targeted therapies in human, in the frame of genetic interventions and personalized medicine. Similarly, to replacing a screw in a machine, in order to figure out how it works, by inducing artificial mutations in mice or other models, helps in deciphering their molecular interactions and cellular mechanisms. Even though mice and transgenic models have been being widely used to address the structural interactions and signalling mechanisms of numerous biological processes, the fact that these models are artificially altered to target /study the underlying mechanisms, there are certain factors that remain questionable. The elephant comes to the rescue as it is a rare fortunate event to discover a naturally living system – in this case the elephant – which naturally employs a regulation mechanism to overcome diseases and environmental stresses; that can be used as a robust living model to inspire research directions. On another aspect, understanding more about the elephants, is vital for taking efficient, evidence-based actions favoring their living conditions regarding environmental / climatic changes (temperature) and pollution, which constitute genotoxic agents.
Corresponding author Dr. Konstantinos Karakostis from Autonomous University of Barcelona, comments “Conceptually, the accumulation of structurally modified p53 pools, collectively or synergistically co-regulating the responses to diverse stresses in the cell, establishes an alternative mechanistic model of cell regulation of high potential significance to biomedical applications”.
Co-author Prof. Fritz Vollrath of Oxford University and Save the Elephants adds: “This I intricate and intriguing study demonstrates how much more there is to elephants than impressive size and how important it is that we not only conserve but also study these signature animals in minute detail. After all, their genetics and physiology are all driven by evolutionary history as well as today’s ecology, diet and behavior.”
Contact:
Umesh Kalathiya (umesh.kalathiya@ug.edu.pl)
Konstantinos Karakostis (Konstantinos.Karakostis@uab.cat)
We are very happy to communicate that our last publication has been accepted in Biomolecules – Emergent Role of IFITM1/3 towards Splicing Factor (SRSF1) and Antigen-Presenting Molecule (HLA-B) in Cervical Cancer (https://doi.org/10.3390/biom12081090)
We investigate new protein–protein interactions for IFITM1/3 in the context of cancer that would shed some light on how IFITM1/3 attenuate the expression of targeted proteins such as HLA-B. Our study has identified new proteins associated with IFITMs which support their role in mediating protein expression; a pivotal function that is highly relevant for viral infection and cancer progression. Our data link IFITM1/3 proteins to HLA-B mRNA and SRSF1 and, all together, our results begin to elucidate how IFITM1/3 catalyze the synthesis of target proteins. Our results suggest that IFITM1/3 affect the expression of HLA-B which could impact the presentation and recognition of oncogenic antigens on the cell surface by cytotoxic T cells and, ultimately, limit tumor cell eradication. In addition, IFITMs are widely studied for their role in inhibiting viruses, and multiple studies have associated IFITMs with cancer progression. Therefore, the role of IFITMs in mediating protein abundance is relevant, as it has the potential for regulating the expression of viral and oncogenic proteins.

We are pleased to announce that our research article entitled “DIA-MS proteome analysis of formalin-fixed paraffin-embedded glioblastoma tissues” has been published (https://www.sciencedirect.com/science/article/pii/S0003267022002665).
The new wave of oncology studies across many applications, including its use in molecular classification, oncogenic pathway analysis, drug, and biomarker discovery, and the unraveling of therapy response and resistance mechanisms. In recent years, the analytical performances of Mass spectrometry (MS)-based proteomics technologies have significantly improved. Currently, it is possible to quantify protein levels in samples using different methodologies. The publication presents, quantitative method called data-independent acquisition MS (DIA-MS) to detect and quantify proteins in glioblastoma formalin-fixed paraffin-embedded (FFPE) tissue microdissections. Owing to their long-term stability, inexpensive storage, and rich clinical data, FFPE tissues are indeed the most valuable resource for protein biomarker research. However, challenges with protein extraction from these specimens are extant, and there is a need for workflows that are simple, robust, and easily implementable. Our work shows the benefit of critically assessing chemical buffers before establishing the most effective workflow for proteome analysis of FFPE tissues. We thus established a precise DIA-MS approach, which enabled the detection of over 1700 proteins and quantified more than 1400 proteins. Further, we demonstrate effective quantification of glioblastoma-relevant and immune-related proteins, allowing more profound insight into biological systems. We revealed a patient-specific proteome, and this approach holds great potential for routinizing protein quantification in FFPE tissue samples.

We are pleased to announce that our review entitled “Interferon Gamma, MHC Class I Regulation and Immunotherapy” has been published (https://doi.org/10.33696/Signaling.3.065). The Major histocompatibility complex class I (MHC-I) molecules are crucial elements for tumor-associated antigen recognition and elimination of cancer cells. In this review, we discuss the regulation of MHC-I, the implications of interferon-gamma (IFNγ) signaling enhancing the expression of MHC-I, and their association with immunotherapy. Many patients diagnosed with various types of cancer have inhibited the IFNγ signaling pathway and present anomalies in the MHC-I expression. In this regard, the review additionally explores how deficiencies associated with IFNγ signaling affect MHC-I expression in tumor cells. Furthermore, we stress the importance of gaining molecular knowledge to improve tumor stratification and find complementary IFNγ effectors that could activate the MHC-I expression. Altogether, it will help predict which tumors can recover expression of MHC-I and respond to immunotherapy.

Since the beginning of the SARS-CoV2 pandemic the ICCVS team has been focused on understanding of the virus recognition mechanisms, immune response and new drug discovery.Recently, prof. Natalia Marek- Trzonkowska, MSCs Łukasz Arcimowicz and VMD Ines Papak from Cancer Immunology group of ICCVS have published a paper on this topic in collaboration with Department of Oncology and Radiotherapy, Department of Infectious Diseases and Department of Pneumonology of the Medical University of Gdańsk.

The paper
Successful Treatment of Persistent SARS-CoV-2 Infection in a B-Cell Depleted Patient with Activated Cytotoxic T and NK Cells: A Case Report Jacek Jassem *, Natalia Maria Marek-Trzonkowska *, Tomasz Smiatacz, Łukasz Arcimowicz, Ines Papak, Ewa Jassem, Jan Maciej Zaucha. Int J Mol Sci. 2021 Oct 10;22(20):10934. doi: 10.3390/ijms222010934.
has been published in the International Journal of Molecular Sciences (IF=5.923): https://www.mdpi.com/1422-0067/22/20/10934.
The publication presents a clinical and immunologic aspects of an immunocompromised lymphoma patient with persistent SARS-CoV-2 viremia successfully treated with a combination of remdesivir and convalescent plasma three months after primary infection. The unique feature of the study is a deep immune profiling of the patient with the focus on the clonal expansion of Th and Tc cells, and the activation status of T and NK cells. Despite of B cell depletion the patient showed robust proliferation and activation of TCR Vβ2 CD8+ T-cell clones and CD16+CD56- natural killer cells during resolution of the infection. The report indicates that SARS-CoV-2 patients with B cell deficiency may achieve an effective clinical response when NK and antigen-specific Tc-cell activation is induced. This is the first study demonstrating TCR-specific oligoclonal response of T cells in B cell deficient patient who recovered from COVID19.
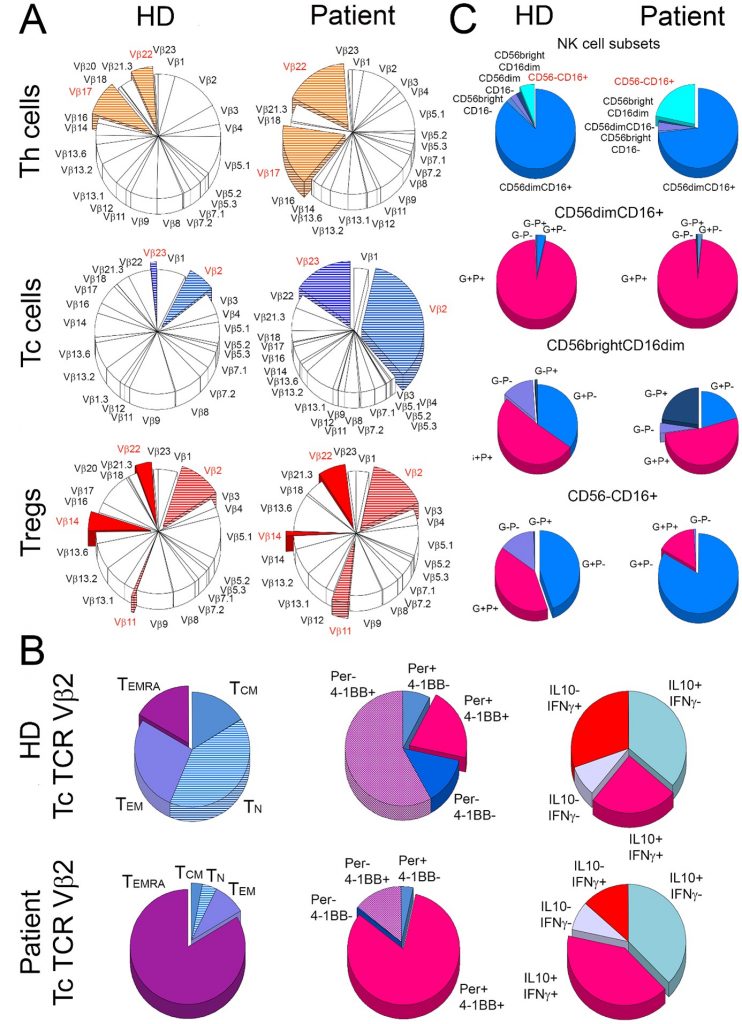
Oligoclonal expansion and activation of T cells in the B-cell depleted patient. The figure depicts (A) mean frequency distribution of 24 T-cell receptor (TCR) Vβ families of T cells in three healthy donors (HD) and the patient (Patient). The most striking differences between the patient and the control donors are marked with colours. (B) shows characteristics of cytotoxic T (Tc) cells from TCR Vβ2 family in healthy donors (HD) and the patient (Patient). For both the patient and the control group, the frequency of naive (TN), central memory (TCM), effector memory (TEM), and effector memory Tc cells that re-expressed CD45RA (TEMRA) were compared (1st column). In addition, TCR Vβ2 Tc cells were compared for the proportions of perforin (Per) and 4-1BB (activation marker) positive (+) and negative (-) cells (2nd column) and for the distribution of cells positive (+) and negative (-) for IL-10 and IFN- (3rd column). The right panel (C) shows the distribution of five subsets of natural killer (NK) cells (1st row) and then cells positive (+) and negative (-) for perforin (P) and granzyme A (G) within CD56dimCD16+ (2nd row), CD56brightCD16dim (3rd row), and CD56-CD16+ (4th row) NK-cell subsets in the patient and healthy donors.
The TAP transporters constitute functionally key components of the antigen presentation pathway since they link the cytosolic pool of peptides with the PLC and the ER-resident MHC-I molecules. The TAP1-TAP2 complex transports antigenic peptide substrates into the endoplasmic reticulum (ER). In ER, the peptides are further processed and loaded on the major histocompatibility class (MHC) I molecules by the peptide loading complex (PLC). The TAP transporters are linked with the PLC; a target for cancers and viral immune evasion. But the mechanisms whereby the cancer-derived mutations in TAP1-TAP2 or viral factors targeting the PLC, interfere peptide transport are only emerging.
on the major histocompatibility class (MHC) I molecules by the peptide loading complex (PLC). The TAP transporters are linked with the PLC; a target for cancers and viral immune evasion. But the mechanisms whereby the cancer-derived mutations in TAP1-TAP2 or viral factors targeting the PLC, interfere peptide transport are only emerging.
This recent study published by M. Padariya et al. describes that transit of peptides through TAP can take place via two different channels (4 or 8 helices) depending on peptide length and sequence. Molecular dynamics and binding affinity predictions of peptide-transporters demonstrated that smaller peptides (8–10 mers; e.g. AAGIGILTV, SIINFEKL) can transport quickly through the transport tunnel compared to longer peptides (15-mer; e.g. ENPVVHFFKNIVTPR). In line with a regulated and selective peptide transport by TAPs, the immunopeptidome upon IFN-γ treatment in melanoma cells induced the shorter length (9-mer) peptide presentation over MHC-I that exhibit a relatively weak binding affinity with TAP. A conserved distance between N and C terminus residues of the studied peptides in the transport tunnel were reported. Furthermore, by adversely interacting with the TAP transport passage or affecting NBD domains tilt movement, the viral proteins and cancer-derived mutations in TAP1-TAP2 may induce allosteric effects in TAP that block conformation of the tunnel (closed towards ER lumen). Interestingly, some cancer-associated mutations (e.g. TAP1R372Q and TAP2R373H) can specifically interfere with selective transport channels (i.e. for longer-peptides). These results provide a model for how viruses and cancer-associated mutations targeting TAP interfaces can affect MHC-I antigen presentation, and how the IFN-γ pathway alters MHC-I antigen presentation via the kinetics of peptide transport.
Movies describes the wild-type TAP1-TAP2 transporters allows the peptide passages through the channel, whereas the mutated TAP complex may hinder the peptide transport process.
A recently published article in Nature Methods entitled „The emerging landscape of single-molecule protein sequencing technologies” jointly led by researchers including dr Javier Alfaro from the ICCVS, University of Gdansk and dr Adam Pomorski from the University of Wrocław, was developed in cooperation with scientists from around the world including significant contributions by dr Umesh Kalathiya, MSc Georges Bedran and prof. David Goodlett from ICCVS. Its aim is to describe the technologies developed for studying proteins at the level of a single molecule, which would enable the determination of the proteome at the level of one cell.
Single-cell sequencing methods are a new technology that allows us to study individual cells within a tissue. In cancer this is helping us to understand why tumours sometimes cannot be treated. While genomes (DNA) and transcriptomes (RNA) can be explored at the single-cell level, single-cell profiling of proteomes (proteins) has been very difficult. Since proteins carry out many of the most important activities in the cell, this gap in technology is greatly limiting our understanding of tissue biology.
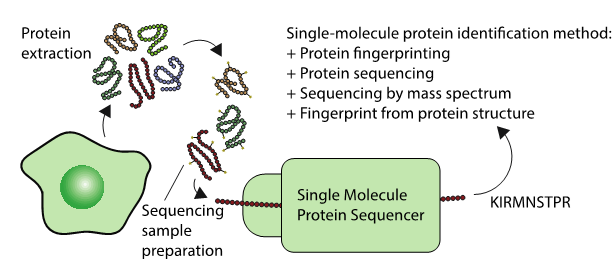
These up and coming sequencing techniques could one day sequence the proteomes of single cells, or lead to new highly sensitive diagnostics. Generally, the techniques involve protein extraction, followed by some sample preparation specific to a diversity of technologies. Each technologies identifies a protein in a different way as described in the figure.
“Proteomics is currently dominated by mass-spectrometry technologies” – says dr Javier Alfaro from the International Center for Cancer Vaccine Science in Gdansk. These technologies are expensive and sequence proteins poorly in complex mixtures. “We do not sequence every amino-acid in the sequence as we do in DNA in these complex mixtures” – says dr Alfaro. The field is really waiting for a transformative wave of technologies similar to what hit the DNA sequencing world – but applied to proteins. “The problem is that unlike DNA, there is no way to amplify proteins – and this poses a big challenge to the sensitivity of current instrumentation” – adds dr Alfaro.
Proteomics awaits a similar transformative wave of protein-sequencing techniques that will allow for the examination of proteins at the single-cell and ultimately single-molecule levels, even with low-abundant proteins. Such techniques would allow routine proteome profiling, like today’s single-cell RNA sequencing studies, creating opportunities for single-cell proteomics and potentially permitting real-time testing for on-site medical diagnostics and disease screening.
“These new technologies depend on new chemistries and biological reporters that allow us to fluorescently detect not only specific amino acids in proteins, but also their modifications” – says dr Adam Pomorski from the University of Wrocław.
During the writing of the work, a blossoming world of biophysicists, technologists, clinicians and computational researchers was invited to share their perspectives as equal contributors. “In this day and age, we are all masters of our own specific fields. Bringing people together as equals so that their full energy and diverse perspectives can be combined is essential to break barriers and bring about scientific insight” – says dr Javier Alfaro.
Read the article: https://www.nature.com/articles/s41592-021-01143-1
Coronavirus disease 2019 (Covid-19) is an infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The 2019-2020 coronavirus outbreak, declared a pandemic by the World Health Organisation on 11 March 2020, caused multiple deaths and changed the way we live and work. There is a huge ongoing international effort to contain the disease and to find effective medications. Many Polish clinicians and scientists are on the front line of the Covid-19-related developments by helping patients in hospital words, performing diagnostic testing, providing advice to policy makers and doing valuable research that could speed up development of effective vaccines and therapeutics.
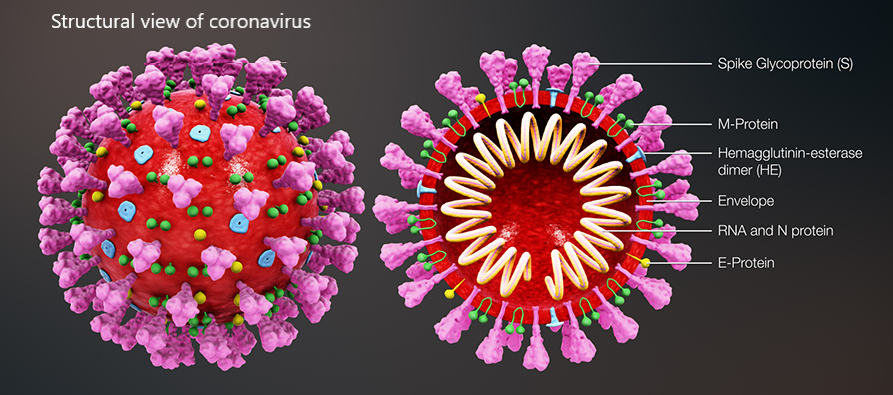
A recent example of Covid-19-centred research contribution from our investigators is a study titled “Highly conserved homotrimer cavity formed by the SARS-CoV-2 spike glycoprotein: a novel binding site”, which has been published in the Journal of Clinical Medicine. This collaborative work has emerged from the International Centre for Cancer Vaccine Science , a joint research unit developed between the University of Gdansk in Poland and the University of Edinburgh co-financed by the Foundation for Polish Science and the European Union Regional Development Fund. The SARS-CoV-2 spike protein plays an important role in the life cycle of the virus by binding to angiotensin converting enzyme-2 (ACE2) receptor on host cells (facilitating entry into the host cell. The team measured spike protein variability derived from 791 viral genomes and studied its properties by molecular dynamics simulation. The molecular docking data revealed a previously unidentified cavity formed by trimerization of the Spike protein that could accommodate macrolide antibiotics such as Rapamycin. The finding of a novel “druggable” site on the Spike protein protein will assist in future drug discovery programs aimed at targeting the coronavirus (CoV) family of viruses using macrocyclic immunomodulatory compounds. This collaboration reinforces the value that the International Research Agenda Programme of the FNP and the Gdańsk University place on working with our international colleagues and emphasizes the need to develop ever stronger international research ties to tackle emerging global health problems.
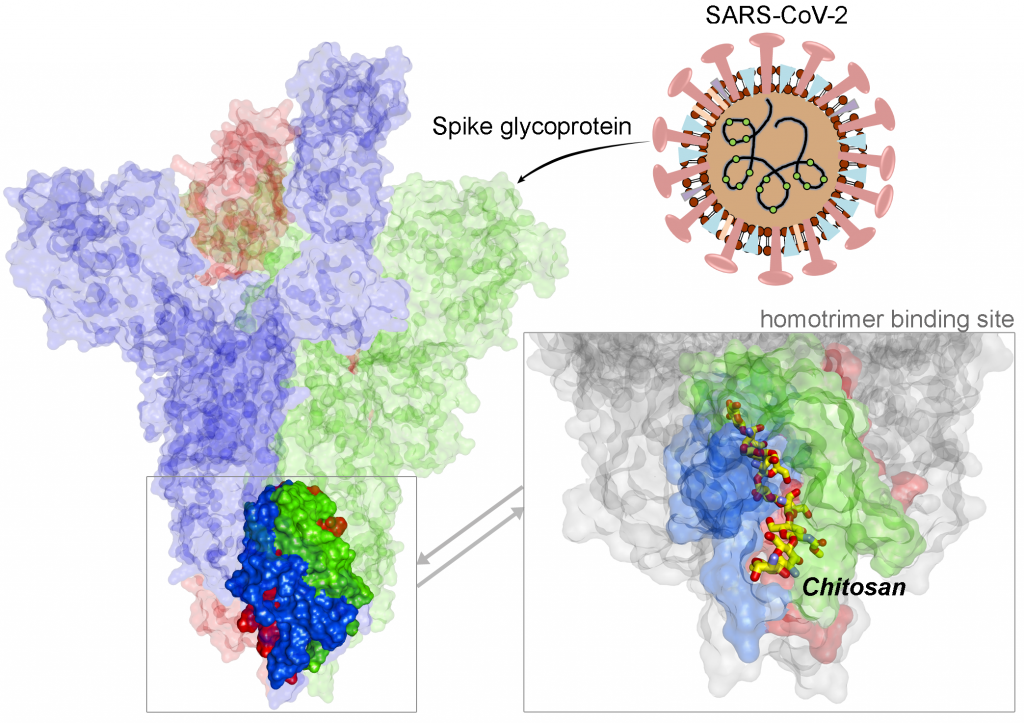
Structural view of SARS-CoV-2 spike protein and location of the Chitosan drug in the identified binding site
Related Links:
- Article in the Journal of Clinical Medicine
- Covid-19 homepage (gov.pl)
- Covid-19 homepage (gov.uk)
- Covid-19 homepage (nhs.uk)
- ICCVS team
- Other publications of ICCVS
Related Stories:
- The first Polish genetic sequence of SARS-CoV-2 coronavirus extracted from the Polish patient
- Innovative University of Gdańsk and BioVentures Institute technology to fight against COVID-19
- First Covid-19 walk-thru mobile testing center in Poland opened at the University of Gdańsk
- IGMM to support NHS by testing Covid-19 samples
- Clinical Fellow Acts as Scotland Lead for UK Coronavirus Cancer Monitoring Programme


