 The National Science Center (NCN) has announced the results of the competition for research projects for young scientists SONATINA 4 Among 35 scientists there is dr inż. Umesh Kalathiya from the International Centre for Cancer Vaccine Science (ICCVS), University of Gdańsk. The project: “Specificity in detection of PTCs in mRNA by NMD and its network, insights from cancer perspective and cross-linking (XL-MS)” received funding in the amount of PLN 604,075.00.
The National Science Center (NCN) has announced the results of the competition for research projects for young scientists SONATINA 4 Among 35 scientists there is dr inż. Umesh Kalathiya from the International Centre for Cancer Vaccine Science (ICCVS), University of Gdańsk. The project: “Specificity in detection of PTCs in mRNA by NMD and its network, insights from cancer perspective and cross-linking (XL-MS)” received funding in the amount of PLN 604,075.00.
The aim of the SONATINA competition is to support the careers of young researchers by enabling full-time employment and research in Poland, as well as gaining knowledge and experience during internships in high-quality foreign research centers.
Congratulations!
More information: www.ncn.gov.pl
Project Summary
Nonsense mediated mRNA decay (NMD) is a quality-control checkpoint that detects and eliminates aberrant messenger RNAs (mRNAs) with premature termination codons (PTCs). The PTCs are often more deleterious compared to missense mutations, as they result in the dysfunctional or complete loss of protein function or expression. In addition, the PTCs arise from single nucleotide mutations that convert a canonical triplet nucleotide codon into one of three stop codons, e.g., TAG, TGA, or TAA. The abundance of mRNA containing the PTCs are reduced through the NMD, but in some cases, the truncated proteins may have dominant negative functions. As such, PTCs represent a unique constellation of diseases which afflict over 30 million people worldwide, accounting for 10-15% of all genetic diseases. Majority of the gene mutations found in genetic disorders, including cancer, result in premature termination codons, and the rapid degradation of their mRNAs by NMD. The nonsense-mediated mRNA decay pathway plays an important role in the degradation of mRNA containing PTCs, but the molecular mechanism and structural rearrangements during this process has not been fully delineated. Genetic disease and cancer represents a significant burden to the society, and any new information, mechanistic details, and molecular properties about different components from NMD machinery could help to bring new therapeutic strategies that are beneficial to the society. The current project originates from an unmet need to understand the NMD mechanisms, and therefore protein-protein or protein-RNA or RNA-RNA interactions will be studied using different in silico (molecular modeling and molecular dyna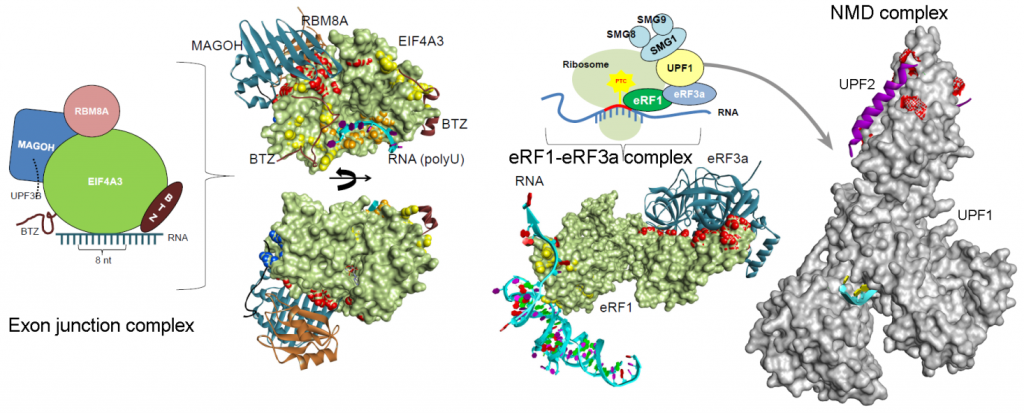 mics) and mass spectrometry (MS)-based structural (cross-linking; XL-MS) techniques (or the proteomics techniques). In addition to the nonsense-mediated mRNA decay pathway, components from eRFs, NMD, and EJC are widely involved in the regulation of telomere maintenance, regulation of chromosome organization, and RNA transport/localization/catabolic process. Therefore, it becomes crucial to understand the properties and effects of cancer mutations on the functioning of different components from the NMD machinery. The outcome of this project will bring a mechanistic overview with characterising molecular properties of the eRFs, NMD, and EJC complexes. Reaching these aims, the project will develop data science techniques aimed at the high-throughput analysis of protein complexes, their structures and methods to put these into quickly use these features for the interpretation of cancer mutation landscapes. Particularly, in the context of RNA-protein interactions this is lacking, adding a novel dimension to structural informatics.
mics) and mass spectrometry (MS)-based structural (cross-linking; XL-MS) techniques (or the proteomics techniques). In addition to the nonsense-mediated mRNA decay pathway, components from eRFs, NMD, and EJC are widely involved in the regulation of telomere maintenance, regulation of chromosome organization, and RNA transport/localization/catabolic process. Therefore, it becomes crucial to understand the properties and effects of cancer mutations on the functioning of different components from the NMD machinery. The outcome of this project will bring a mechanistic overview with characterising molecular properties of the eRFs, NMD, and EJC complexes. Reaching these aims, the project will develop data science techniques aimed at the high-throughput analysis of protein complexes, their structures and methods to put these into quickly use these features for the interpretation of cancer mutation landscapes. Particularly, in the context of RNA-protein interactions this is lacking, adding a novel dimension to structural informatics.


